Introduction¶
ChemTreeMap is a novel program for visualization of biochemical similarity in molecular datasets. It allows for users to interactively explore the chemical space covered by a set of small molecules. It is written by Jing Lu, a PhD student under the supervision of Dr. Heather Carlson.
Software¶
Briefly, the data processing is done using Python for calculations of similarity, and JavaScript for visualization:
The workflow is shown in the following Fig. 1.1.
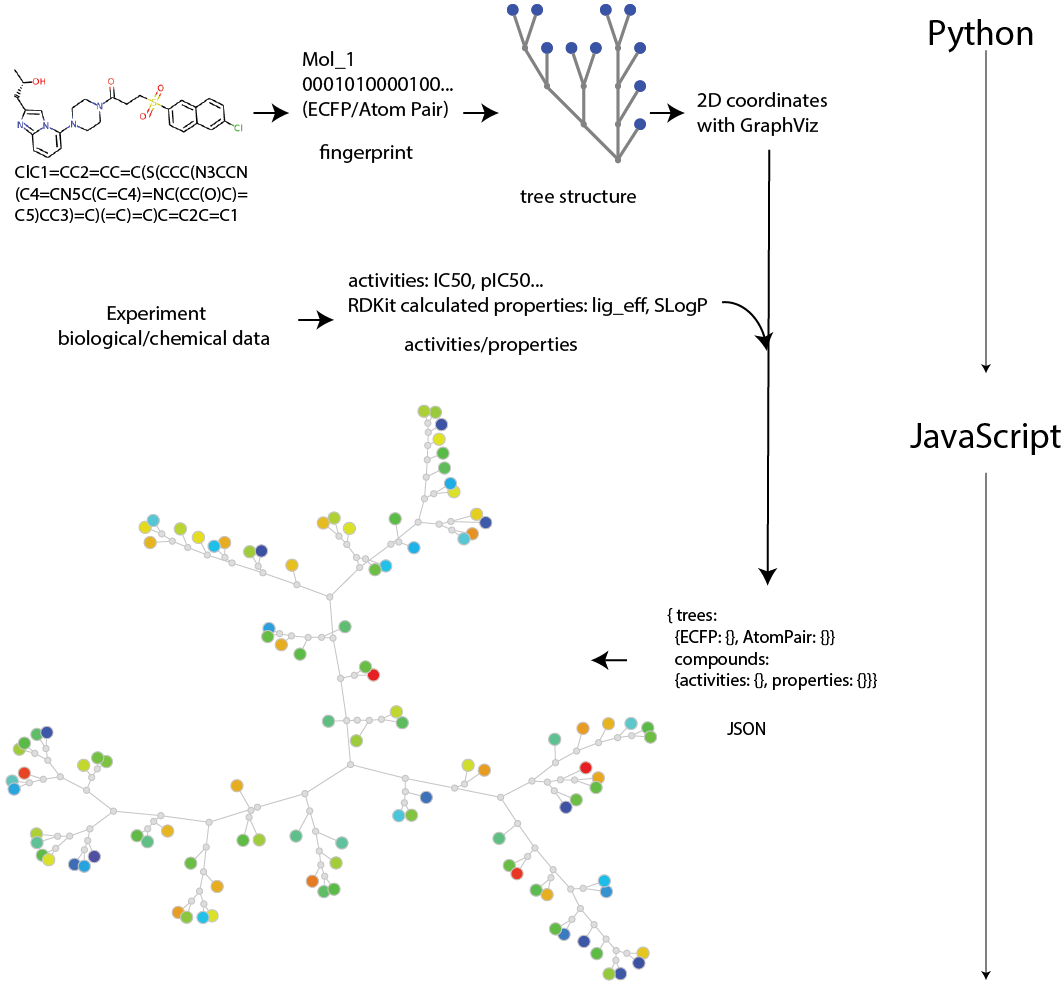
The workflow for ChemTreeMap
There is a demo (http://ajing.github.io/ChemTreeMap/index.html#example) of the interactive frontend for some example datasets hosted on GitHub.
Paper¶
ChemTreeMap: An Interactive Map of Biochemical Similarity in Molecular Datasets.
The paper has been submitted to Bioinformatics.